
Genomics of Pediatric Cancers
Pediatric cancers often have few targetable mutations in cancer genes, even in cases that are very advanced. In cases where gene alterations are found, these are often fusion genes or other alterations not easily druggable. We use integrative genomics to identify new therapeutic opportunities for advanced pediatric cancers. We have extensive experience in generating and using next-generation sequencing data for gene and network discovery.
Our laboratory has sequenced over 300 pediatric tumors by whole-genome (WGS) and RNA sequencing (RNAseq), as part of a multidisciplinary effort to apply next-generation sequencing to advance the care of relapsed and other high-risk pediatric cancer patients at UCSF/Benioff Children’s Hospitals. We have also generated over 50 PDX models and a number of PDX-derived cell lines which we use to study aspects of cancer biology. Our work is also facilitated by ongoing and intensive tumor banking efforts at UCSF Benioff Children’s Hospitals.
Key efforts involve identifying possible actionable alterations, analyzing longitudinal samples, studying cancer evolution and incorporating advanced genomic tools including mutational signature analysis and in silico assessment of heterogeneity in the tumor microenvironment. We have recently expanded our “bulk” sequencing to performing single-cell and spatially resolved sequencing.
Our extensive sequencing datasets provide rich research opportunities for post-docs and students interested in the intersection of cancer biology, functional genomics, and computational biology. Pediatric cancer cases are frequently discussed in a multidisciplinary molecular tumor board which all trainees are encouraged to attend, providing a “front row” to the clinical application of genomic analysis in clinical care.
Projects
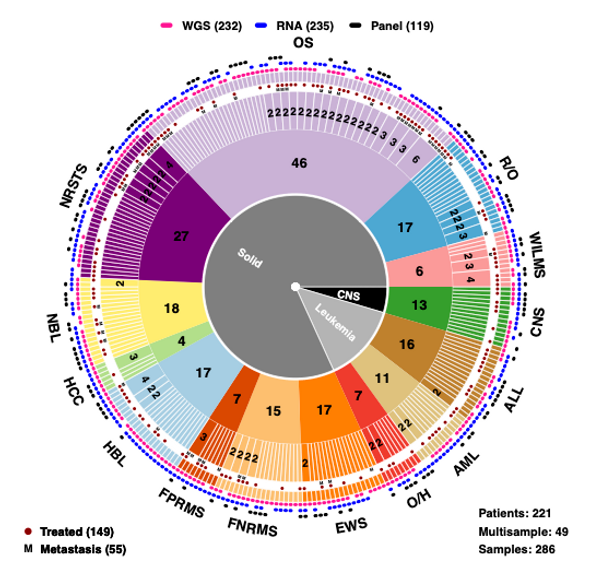
Longitudinal profiling of high-risk pediatric malignancies using a multiomics approach
For many pediatric cancer patients, commonly used gene-panel sequencing tests yield few actionable results, partly due to the complex genomic alterations present. We hypothesized that an unbiased sequencing methods including, WGS and RNAseq and other omic appraches, can overcome this and lead to a more comprehensive understanding of these diseases. While prior studies have evaluated WGS and RNAseq in pediatric cancers, few focused primarily on metastatic or relapsed disease. We have placed special focus on longitudinal profiling of patients, including with additional deep sequencing, to capture tumor evolution at the primary and metastatic sites, and to quantify the utility of resampling.
High-resolution mapping of oncogenic structural changes in osteosarcoma
We have leveraged long-read and spatial sequencing modalities to identify cancer-specific oncogenic alterations for treatment of osteosarcomas (OS). OS is archetypal of cancers driven by aneuploidy and structural rearrangement rather than point mutations. The functional consequences of many SVs stem from their effect on multidimensional genome organization. SVs can enable enhancer hijacking, alter boundaries of topologically associated domains (TADs), and move gene loci to different regulatory compartments to affect gene expression. SVs can also lead to the formation of extrachromosomal DNA amplicons (ecDNA), which can be megabases in size and can incorporate both genes and regulatory elements to result in substantial increases in gene transcription and intratumor heterogeneity. Complex SVs may have profound implications for prognosis and treatment in OS and other cancers, yet they remain poorly understood. By incorporating long-read optical genome mapping (OGM) and chromatin conformation capture sequencing (HiC), we study SVs in OS with the goal of elucidating their contribution to cancer development.


UCSF500
The discovery-focused efforts of our laboratory dovetail well with an extensive and well-developed clinical precision cancer medicine program. A key to this program is the UCSF500 sequencing assay, a CLIA-certified molecular assay that can identify mutations, fusion, and copy number changes in clinical samples. Unlike many commercially available assays, this test explicitly contains the most important alterations relevant to pediatric solid tumors. Over 3,000 pediatric cancer cases have been sequenced at UCSF using this assay. Much of this data is available externally through our participation in the AACR GENIE consortium. As an internal resource, extensive clinical data extracted from the electronic health record is also available for these cases; this provides an additional invaluable tool for identifying novel cancer drivers in pediatric cancer.
The Sweet-Cordero Lab
University of California, San Francisco
Dept. of Pediatrics
1550 4th Street
Rock Hall Building, Room 382
San Francisco, CA 94158
Leanne Sayles
Laboratory Manager
Email: Leanne.Sayles@ucsf.edu
Flora Ignacio
Admin Assistant
Email: Flora.Ignacio@ucsf.edu