
Pre-clinical Therapeutics
A key problem for advancing OS research is that this is a rare disease and primary tumor samples from patients are difficult to obtain.
We have established a large (over 30 to date) bank of primary xenograft samples from both pre- and post-chemotherapy patient samples from OS patients. We have also generated a panel of cell lines from these PDX models. We have used these PDX models and matching tumors to identify potential targeted therapies for subsets of OS patients (Sayles and Breese et al, Cancer Discovery 2019). This work has had significant impact in the field as previously it was not widely appreciated that OS could be vulnerable to targeted therapies directs at highly amplified (but not mutated) cancer genes such as MYC, CDK4 and others.
OS is characterized by structural rearrangements and copy number changes which drive considerable heterogeneity. What is less clear is how these genetic rearrangements alter are in turn regulated at the epigenetic level to drive gene expression. Defining the epigenetic circuitry of OS could help to understand the role of chromatin accessibility in determining cell plasticity. Chromatin accessibility can prime gene expression and define a cellular state. As chromatin accessibility is a major determinant of epigenetic transcriptional regulation, understanding the variability of chromatin accessibility could identify clinically-relevant subtypes and biomarkers of therapy response in OS, a disease that has seen few advances in therapy for over 40 years.
We are especially interested in using PDX models and genetically engineered models of OS to study metastasis. We have ongoing interest in testing new therapies for osteosarcoma treatment, especially for metastatic disease.
Projects
Evaluating the DNA damage response in osteosarcoma
Current treatment strategies for osteosarcoma rely on highly toxic chemotherapy that has not changed in over forty years. Any advances in therapy will likely require the use of novel, rational approaches for combination therapies. As such, we developed a panel of patient-derived xenograft cell lines (PDXC) obtained from patients at diagnosis, resection, and metastasis that reflect the genotypic heterogeneity seen in patients. Using this panel, single agent and combination drug screens were performed. Combining gemcitabine with agents targeting the ATR-Chk1-Wee1 pathway resulted in drug synergy, an increase in DNA damage and replication stress. Validation using an orthotopic to metastatic model showed that combining gemcitabine and ATRi effectively reduced the primary tumor volume, resulted in a long-lasting decrease in metastatic lesions, and increased survival. These findings provide a strong rationale for the further development of clinical trials involving the inhibition of ATR-Chk1-Wee1 in combination with gemcitabine for the treatment of osteosarcoma.

Defining epigenetic subtypes of osteosarcoma
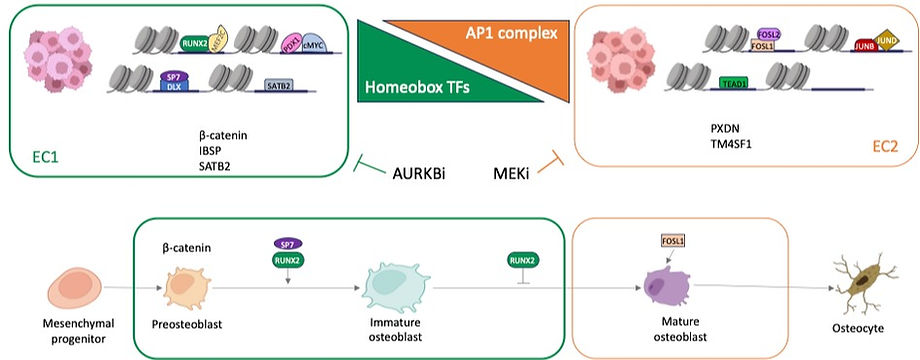
Using ATAC-seq, we identified two epigenetic subtypes (EC1 and EC2) of OS with distinct chromatin accessibility. These studies were done in a panel of 11 PDX-derived cell lines and 9 established OS cell lines. We also performed ATACseq on the matching PDX for the 11 PDX-derived lines as well as 8 primary tumor samples. Analysis of transcription factor (TF) motifs and footprints of ATAC-seq peaks identified transcription factors distinctly activated in these two subtypes. For example, EC1 is characterized by high level of activity of developmental transcription factors including RUNX2, MEF2C and homeobox TFs. In contrast, EC2 is characterized by high activity of AP1 transcription factors such as FOSL1 and FOSL2. In order to understand the underlying transcriptional circuitry of these cell states, we performed CUT&RUN assay for the complete panel of PDX-derived cell lines. We found that the subtypes are driven by enhancer regulation (H3K27ac). In parallel, we used our PDX-derived cell line panel to screen cluster-specific targeted therapies and found differential responses dependent on cellular states defined by chromatin accessibility. For example, cell lines in the EC2 cluster are marked by upregulation of AP1 transcription factors, have evidence of ERK activation and are highly responsive to ERK inhibitors. In contrast cell lines in the EC1 cluster are sensitive to AURKA and AURKB inhibitors. Single-cell RNAseq analysis indicates that these subtypes may coexist within a single tumor. In summary, we discovered two epigenetically distinct cell states that are controlled by a state-specific collection of transcription factors, creating a gene signature that may aid in distinguishing osteosarcoma subclasses and predicting treatment response.
The Sweet-Cordero Lab
University of California, San Francisco
Dept. of Pediatrics
1550 4th Street
Rock Hall Building, Room 382
San Francisco, CA 94158
Leanne Sayles
Laboratory Manager
Email: Leanne.Sayles@ucsf.edu
Flora Ignacio
Admin Assistant
Email: Flora.Ignacio@ucsf.edu